Achieving equalized odds on synthetic data
NOTE: this notebook has extra requirements, install them with:
pip install "error_parity[dev]"
[1]:
import logging
from itertools import product
import numpy as np
import cvxpy as cp
from scipy.spatial import ConvexHull
from sklearn.metrics import roc_curve
[2]:
from error_parity import __version__
print(f"Notebook ran using `error-parity=={__version__}`")
Notebook ran using `error-parity==0.3.11`
Given some data (X, Y, S)
[3]:
def generate_synthetic_data(n_samples: int, n_groups: int, prevalence: float, seed: int):
"""Helper to generate synthetic features/labels/predictions."""
# Construct numpy rng
rng = np.random.default_rng(seed)
# Different levels of gaussian noise per group (to induce some inequality in error rates)
group_noise = [0.1 + 0.3 * rng.random() / (1+idx) for idx in range(n_groups)]
# Generate predictions
assert 0 < prevalence < 1
y_score = rng.random(size=n_samples)
# Generate labels
# - define which samples belong to each group
# - add different noise levels for each group
group = rng.integers(low=0, high=n_groups, size=n_samples)
y_true = np.zeros(n_samples)
for i in range(n_groups):
group_filter = group == i
y_true_groupwise = ((
y_score[group_filter] +
rng.normal(size=np.sum(group_filter), scale=group_noise[i])
) > (1-prevalence)).astype(int)
y_true[group_filter] = y_true_groupwise
### Generate features: just use the sample index
# As we already have the y_scores, we can construct the features X
# as the index of each sample, so we can construct a classifier that
# simply maps this index to our pre-generated predictions for this clf.
X = np.arange(len(y_true)).reshape((-1, 1))
return X, y_true, y_score, group
[4]:
# N_GROUPS = 4
N_GROUPS = 3
# N_SAMPLES = 1_000_000
N_SAMPLES = 100_000
SEED = 23
X, y_true, y_score, group = generate_synthetic_data(
n_samples=N_SAMPLES,
n_groups=N_GROUPS,
prevalence=0.25,
seed=SEED)
[5]:
actual_prevalence = np.sum(y_true) / len(y_true)
print(f"Actual global prevalence: {actual_prevalence:.1%}")
Actual global prevalence: 26.6%
[6]:
EPSILON_TOLERANCE = 0.05
FALSE_POS_COST = 1
FALSE_NEG_COST = 1
# L_P_NORM = np.inf
L_P_NORM = 2
Given a trained predictor (that outputs real-valued scores)
[7]:
# Example predictor that predicts the synthetically produced scores above
predictor = lambda idx: y_score[idx]
Construct the fair optimal classifier (derived from the given predictor)
Fairness is measured by the equal odds constraint (equal FPR and TPR among groups);
optionally, this constraint can be relaxed by some small tolerance;
Optimality is measured as minimizing the expected loss,
parameterized by the given cost of false positive and false negative errors;
[8]:
from error_parity import RelaxedThresholdOptimizer
postproc_clf = RelaxedThresholdOptimizer(
predictor=predictor,
constraint="equalized_odds",
tolerance=EPSILON_TOLERANCE,
false_pos_cost=FALSE_POS_COST,
false_neg_cost=FALSE_NEG_COST,
l_p_norm=L_P_NORM,
max_roc_ticks=None,
seed=SEED,
)
[9]:
%%time
import logging
logging.basicConfig(level=logging.INFO, force=True)
postproc_clf.fit(X=X, y=y_true, group=group)
INFO:root:ROC convex hull contains 0.8% of the original points.
INFO:root:ROC convex hull contains 1.1% of the original points.
INFO:root:ROC convex hull contains 1.7% of the original points.
INFO:root:cvxpy solver took 0.000574791s; status is optimal.
INFO:root:Optimal solution value: 0.17475045311134396
INFO:root:Variable Global ROC point: value [0.07436224 0.54775546]
INFO:root:Variable ROC point for group 0: value [0.10639373 0.53750647]
INFO:root:Variable ROC point for group 1: value [0.05919955 0.55402037]
INFO:root:Variable ROC point for group 2: value [0.05891875 0.55319488]
CPU times: user 199 ms, sys: 8.76 ms, total: 208 ms
Wall time: 1.6 s
[9]:
<error_parity.threshold_optimizer.RelaxedThresholdOptimizer at 0x11df428c0>
Plot solution
[10]:
from matplotlib import pyplot as plt
import seaborn as sns
sns.set(style="whitegrid", rc={'grid.linestyle': ':'})
[11]:
from error_parity.plotting import plot_postprocessing_solution
plot_postprocessing_solution(
postprocessed_clf=postproc_clf,
plot_roc_curves=True,
plot_roc_hulls=True,
dpi=200, figsize=(5, 5),
)
plt.show()
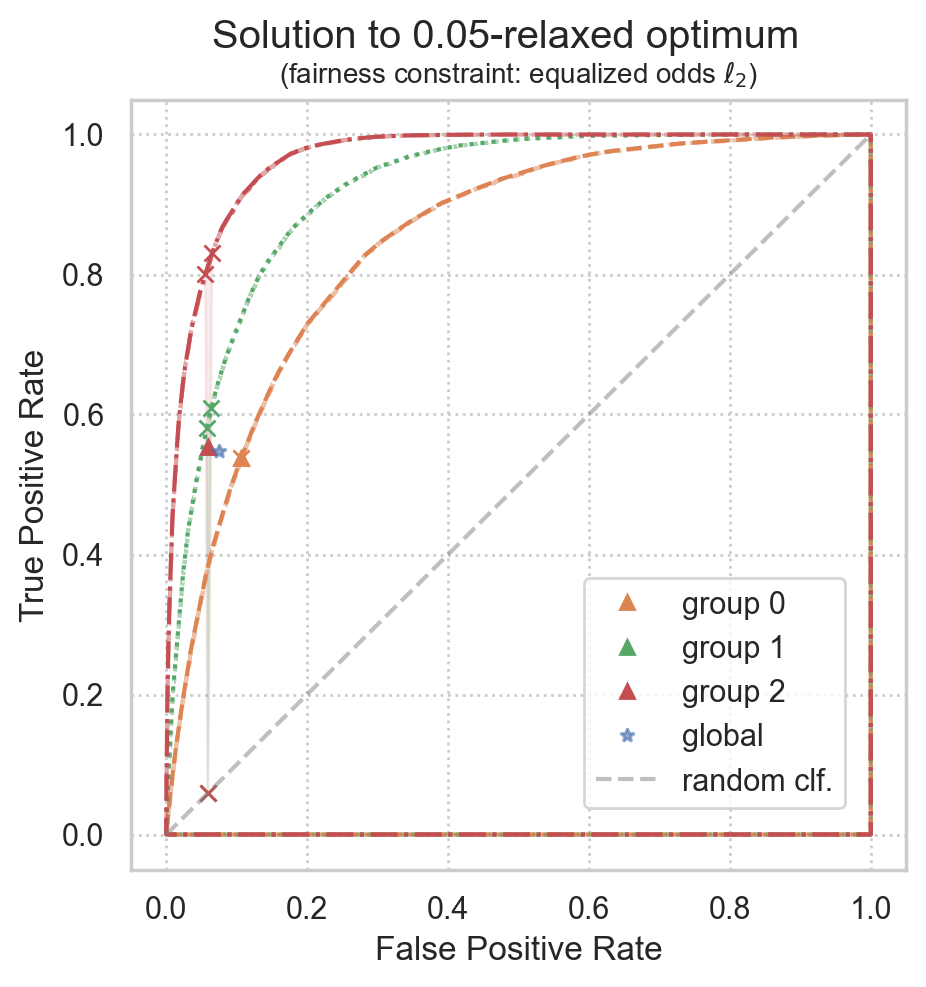
Plot realized ROC points
realized ROC points will converge to the theoretical solution for larger datasets, but some variance is expected for smaller datasets
[12]:
# Set group-wise colors and global color
palette = sns.color_palette(n_colors=N_GROUPS + 1)
global_color = palette[0]
all_group_colors = palette[1:]
[13]:
from error_parity.roc_utils import compute_roc_point_from_predictions
plot_postprocessing_solution(
postprocessed_clf=postproc_clf,
plot_roc_curves=True,
plot_roc_hulls=True,
plot_relaxation=True,
dpi=200, figsize=(5, 5),
)
# Compute predictions
y_pred_binary = postproc_clf(X, group=group)
# Plot the group-wise points found
realized_roc_points = list()
for idx in range(N_GROUPS):
# Evaluate triangulation of target point as a randomized clf
group_filter = group == idx
curr_realized_roc_point = compute_roc_point_from_predictions(y_true[group_filter], y_pred_binary[group_filter])
realized_roc_points.append(curr_realized_roc_point)
plt.plot(
curr_realized_roc_point[0], curr_realized_roc_point[1],
color=all_group_colors[idx],
marker="*", markersize=8,
lw=0,
)
realized_roc_points = np.vstack(realized_roc_points)
# Plot actual global classifier performance
global_clf_realized_roc_point = compute_roc_point_from_predictions(y_true, y_pred_binary)
plt.plot(
global_clf_realized_roc_point[0], global_clf_realized_roc_point[1],
color=global_color,
marker="*", markersize=8,
lw=0,
)
plt.xlim(-0.003, 0.3)
plt.ylim(-0.01, 1.0)
plt.show()
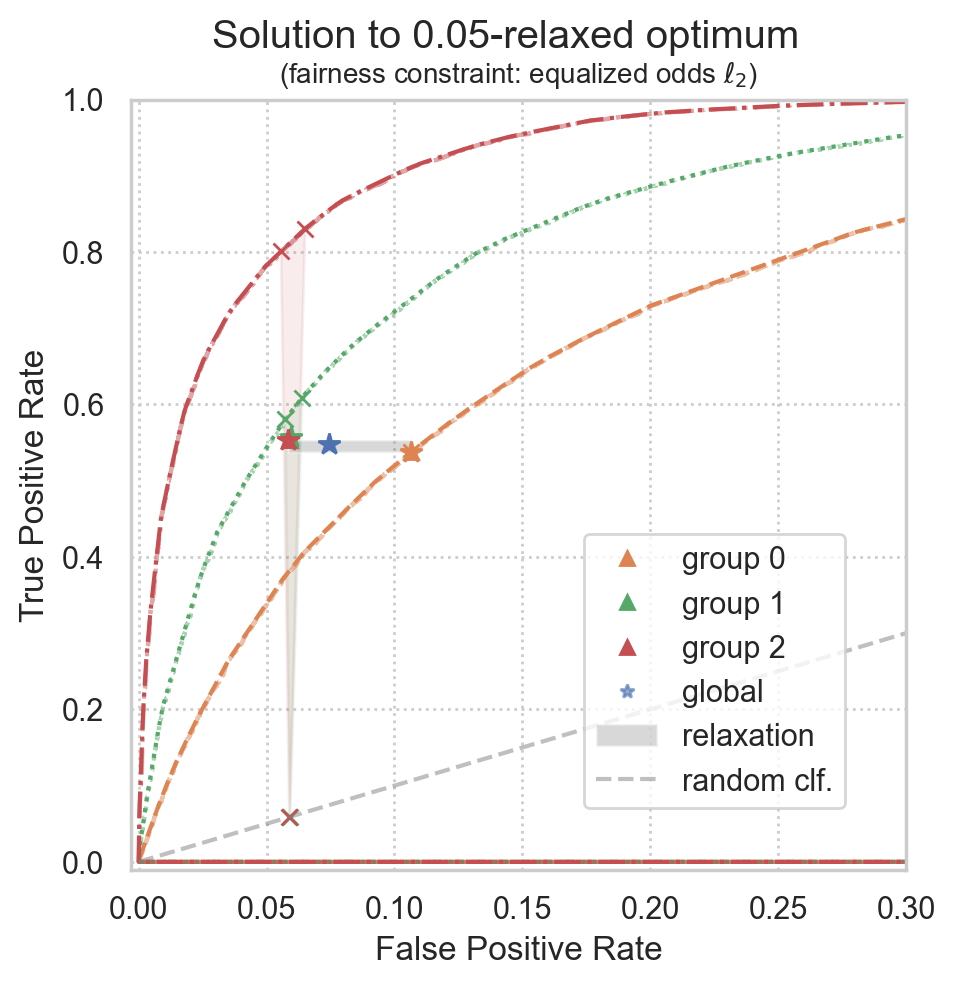
Compute distances between theorized ROC points and empirical ROC points
[14]:
# Distances to group-wise targets:
for i, (target_point, actual_point) in enumerate(zip(postproc_clf.groupwise_roc_points, realized_roc_points)):
dist = np.linalg.norm(target_point - actual_point, ord=2)
print(f"Group {i}: l2 distance from target to realized point := {dist:.3%}")
# Distance to global target point:
dist = np.linalg.norm(postproc_clf.global_roc_point - global_clf_realized_roc_point, ord=2)
print(f"Global l2 distance from target to realized point := {dist:.3%}")
Group 0: l2 distance from target to realized point := 0.000%
Group 1: l2 distance from target to realized point := 0.282%
Group 2: l2 distance from target to realized point := 0.063%
Global l2 distance from target to realized point := 0.089%
Compute performance differences
assumes FP_cost == FN_cost == 1.0
[15]:
from sklearn.metrics import accuracy_score
from error_parity.roc_utils import calc_cost_of_point
# Empirical
accuracy_val = accuracy_score(y_true, y_pred_binary)
# Theoretical
theoretical_global_cost = calc_cost_of_point(
fpr=postproc_clf.global_roc_point[0],
fnr=1 - postproc_clf.global_roc_point[1],
prevalence=y_true.sum() / len(y_true),
)
print(f"Actual accuracy: \t\t\t{accuracy_val:.3%}")
print(f"Actual error rate (1 - Acc.):\t\t{1 - accuracy_val:.3%}")
print(f"Theoretical cost of solution found:\t{theoretical_global_cost:.3%}")
Actual accuracy: 82.559%
Actual error rate (1 - Acc.): 17.441%
Theoretical cost of solution found: 17.475%
[16]:
print(f"Accuracy for dummy constant classifier: {max(np.mean(y_true==label) for label in {0, 1}):.1%}")
Accuracy for dummy constant classifier: 73.4%
[17]:
from error_parity.evaluation import eval_accuracy_and_equalized_odds
lp_acc, lp_eq_odds = eval_accuracy_and_equalized_odds(y_true, y_pred_binary, group, l_p_norm=L_P_NORM)
print(f"Realized LP accuracy:\t {lp_acc:.1%}")
print(f"Realized LP eq. odds violation: {lp_eq_odds:.1%}\n")
Realized LP accuracy: 82.6%
Realized LP eq. odds violation: 5.1%
Plot postprocessing Pareto frontier
i.e., all attainable optimal trade-offs for this predictor
[18]:
from error_parity.pareto_curve import compute_postprocessing_curve
postproc_results_df = compute_postprocessing_curve(
model=predictor,
fit_data=(X, y_true, group),
eval_data={
"fit": (X, y_true, group),
},
fairness_constraint="equalized_odds",
l_p_norm=L_P_NORM,
bootstrap=True,
seed=SEED,
predict_method="__call__",
)
INFO:root:Using `n_jobs=9` to compute adjustment curve.
INFO:root:Computing postprocessing for the following constraint tolerances: [0. 0.01 0.02 0.03 0.04 0.05 0.06 0.07 0.08 0.09 0.1 0.11 0.12 0.13
0.14 0.15 0.16 0.17 0.18 0.19 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 ].
[19]:
from error_parity.plotting import plot_postprocessing_frontier
from matplotlib import pyplot as plt
plot_postprocessing_frontier(
postproc_results_df,
perf_metric="accuracy",
disp_metric=f"equalized_odds_diff_l{postproc_clf.l_p_norm}",
show_data_type="fit",
constant_clf_perf=max((y_true == const_pred).mean() for const_pred in {0, 1}),
model_name=r"$\bigstar$",
)
plt.xlabel(r"accuracy $\rightarrow$")
l_p_norm_str = str(postproc_clf.l_p_norm) if postproc_clf.l_p_norm != np.inf else r"\infty"
plt.ylabel(r"equalized odds $\ell_" + l_p_norm_str + r"$ violation $\leftarrow$")
plt.show()
